 Author
Author  Correspondence author
Correspondence author
Animal Molecular Breeding, 2024, Vol. 14, No. 4
Received: 16 May, 2024 Accepted: 01 Jul., 2024 Published: 17 Jul., 2024
This study investigates the significance of epigenetic markers, such as DNA methylation, histone modifications, and non-coding RNAs, in regulating disease resistance in dogs. A comparative analysis of disease-resistant and susceptible dog breeds highlights key epigenetic differences, shedding light on potential applications for enhancing disease resistance. Despite the technical and ethical challenges associated with using epigenetic data in breeding, the study underscores the long-term potential of epigenetic profiling for disease prevention and health monitoring in dogs. By identifying specific epigenetic markers, this study provides a foundation for improving canine breeding programs and personalized veterinary medicine.
1 Introduction
Disease resistance in dogs is a critical area of veterinary research, as it directly impacts the health and well-being of companion animals. Various pathogens, including bacteria, viruses, and parasites, pose significant threats to canine health, leading to conditions such as antimicrobial resistance and susceptibility to diseases like leishmaniasis and hookworm infections (Tyson et al., 2021). Understanding the genetic and immunological factors that contribute to disease resistance can help in developing effective strategies for disease prevention and management in dogs (Vasconcelos et al., 2017).
Epigenetics, the study of heritable changes in gene expression that do not involve changes to the underlying DNA sequence, plays a crucial role in disease resistance (Wegener et al., 2018). Epigenetic mechanisms, such as DNA methylation, histone modification, and non-coding RNA regulation, can influence how genes are expressed in response to environmental factors, including pathogen exposure (Wang et al., 2023). These modifications can affect the immune response and the ability of dogs to resist infections, making the identification of epigenetic markers a promising area for enhancing disease resistance (Gul et al., 2022).
Identifying epigenetic markers associated with disease resistance in dogs has significant implications for veterinary medicine (Ceric et al., 2019). These markers can serve as biomarkers for early detection of susceptibility to diseases, allowing for timely interventions and personalized treatment plans. Moreover, understanding the epigenetic landscape can lead to the development of novel therapeutic strategies that target specific epigenetic modifications to enhance disease resistance (Islam et al., 2020). This approach can reduce the reliance on antibiotics and other treatments, thereby mitigating the risk of antimicrobial resistance (Mahfouz et al., 2020).
This study identifies and evaluates current knowledge on epigenetic markers of disease resistance in dogs, highlighting the potential of epigenetic research to improve canine health and disease management, including examining genetic and epigenetic factors, the role of specific epigenetic mechanisms, and the implications of these findings for veterinary practice and future research, with the aim of aiding further exploration of epigenetic regulation of disease resistance in dogs and developing targeted health interventions.
2 Epigenetic Mechanisms in Disease Resistance
2.1 DNA methylation
DNA methylation is a crucial epigenetic modification that involves the addition of a methyl group to the DNA molecule, typically at cytosine bases in CpG dinucleotides (Zhou et al., 2020). This process can regulate gene expression by altering the accessibility of the DNA to transcriptional machinery, thereby playing a significant role in genomic stability and disease resistance. For instance, DNA methylation-related long non-coding RNAs (lncRNAs) have been shown to modulate gene expression by interacting with chromosomal modifications or remodeling factors, which can impact the progression of diseases such as lower-grade gliomas (LGGs). Additionally, DNA methylation is a reversible modification, making it a potential target for therapeutic interventions in various diseases (Izquierdo and Crujeiras, 2019).
2.2 Histone modifications
Histone modifications, including acetylation, methylation, phosphorylation, and ubiquitination, are another layer of epigenetic regulation that influences chromatin structure and gene expression. These modifications can either promote or repress transcription depending on the specific type and location of the modification. Histone deacetylation and methylation inhibitors have been approved for clinical use in treating hematological malignancies, highlighting their therapeutic potential (Begolli et al., 2019). Furthermore, lncRNAs have been found to interact with histone-modifying enzymes, thereby influencing histone modifications and contributing to disease resistance mechanisms (Gray et al., 2022).
2.3 Non-coding RNAs (miRNA, lncRNA)
Non-coding RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), play a pivotal role in the epigenetic regulation of gene expression. miRNAs are short non-coding RNAs that can silence gene expression post-transcriptionally, while lncRNAs can interact with chromatin and other epigenetic machinery to regulate gene expression. For example, miRNAs have been shown to be epigenetically regulated by DNA methylation and histone modifications, and their dysregulation is associated with various diseases, including cancers (Pathania et al., 2021). Similarly, lncRNAs are involved in a wide range of biological processes and can modulate gene expression through interactions with epigenetic regulators, contributing to disease resistance (Raei et al., 2021).
2.4 Interaction between epigenetics and genomic regulation
The interaction between epigenetic mechanisms and genomic regulation is complex and involves multiple layers of control. Epigenetic modifications such as DNA methylation, histone modifications, and non-coding RNAs work in concert to regulate gene expression and maintain genomic stability. Disruption in these interactions can lead to disease progression and resistance to therapies. For instance, the crosstalk between lncRNAs and miRNAs has been shown to play a critical role in drug resistance in gastrointestinal cancers, highlighting the importance of understanding these interactions for developing effective therapeutic strategies5. Additionally, the integration of epigenetic and genomic data is essential for identifying novel biomarkers and therapeutic targets for disease resistance (Maimaiti et al., 2022). In summary, the study of epigenetic mechanisms, including DNA methylation, histone modifications, and non-coding RNAs, provides valuable insights into the regulation of gene expression and disease resistance. Understanding the intricate interactions between these mechanisms and genomic regulation is crucial for developing novel therapeutic approaches and improving clinical outcomes (Piletič and Kunej, 2016).
3 Current Research on Epigenetic Markers in Canine Health
3.1 Identified epigenetic markers for disease resistance
Recent studies have identified several epigenetic markers that play a crucial role in disease resistance in dogs. For instance, the epigenetic regulation of the ABCB1 gene has been shown to be significant in determining the multidrug resistance (MDR) phenotype in canine lymphoid tumor cell lines (Tomiyasu et al., 2014). Specifically, differences in DNA methylation and histone H3 acetylation were observed between drug-sensitive and drug-resistant cell lines, indicating that these epigenetic modifications are associated with the MDR phenotype. Additionally, research on canine malignant lymphoma has highlighted the potential of histone deacetylases and demethylase inhibitors as future treatments, as well as the use of microRNAs as diagnostic and prognostic biomarkers (Figure 1) (Montaner-Angoiti et al., 2023).
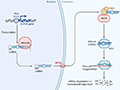 Figure 1 Scheme of miRNA biogenesi (Adopted from Montaner-Angoiti et al., 2023) Image caption: In the nucleus, miRNA is transcribed by RNA polymerase II as primary transcripts (pri-miRNA). The Drosha enzyme cuts this pri-miRNA to form a premiRNA, which is actively transported to the cytoplasm by the nuclear transport receptor exportin 5 (XPO5). In the cytoplasm, the pre-miRNA is cut by a second enzyme, Dicer, to form a mature and short double-stranded miRNA molecule. The miRNA duplex is incorporated into the RISC protein complex (Adopted from Montaner-Angoiti et al., 2023) |
Montaner-Angoiti et al. (2023) explored the critical processes of miRNA biogenesis, shedding light on the sequential stages of miRNA maturation. In the nucleus, miRNA transcription produces primary transcripts (pri-miRNA), which are processed by the Drosha enzyme. This early processing step is essential to convert pri-miRNA into pre-miRNA, a necessary precursor for downstream maturation. Afterward, exportin 5 (XPO5) mediates the active export of pre-miRNA to the cytoplasm. Once in the cytoplasm, the Dicer enzyme further refines pre-miRNA by cleaving it into a mature double-stranded miRNA duplex. Importantly, the mature miRNA is subsequently integrated into the RNA-induced silencing complex (RISC), which is responsible for gene silencing activities such as mRNA degradation or translational repression. This pathway underlines the precise regulatory role of miRNAs in post-transcriptional gene expression, providing potential targets for therapeutic interventions.
3.2 Advances in epigenomic technologies for marker identification
Advances in epigenomic technologies have significantly enhanced the identification of epigenetic markers in canine health. Techniques such as bisulphite sequencing, real-time methylation-specific PCR, and chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) have been employed to study the epigenetic regulation of genes associated with disease resistance. For example, bisulphite sequencing and real-time methylation-specific PCR were used to reveal hypermethylation and hypomethylation patterns in the CpG islands of the ABCB1 gene in different canine lymphoid tumor cell lines (Ling and Rönn, 2016). Furthermore, ChIP-qPCR has been utilized to assess histone modifications, providing insights into the epigenetic landscape of drug-resistant and drug-sensitive cell lines (Izquierdo and Crujeiras, 2019).
3.3 Case studies from other species relevant to canine epigenetics
Research on epigenetic markers in other species offers valuable insights that can be applied to canine health (Furtado et al., 2019). For instance, studies on insulin resistance in humans have demonstrated that epigenetic variations, such as DNA methylation, play a significant role in disease development and progression. These findings suggest that similar epigenetic mechanisms may be involved in canine diseases. Additionally, research on the epigenetic regulation of gene function in cancer has shown that targeting epigenetic alterations can be a promising therapeutic strategy. This concept has been explored in the context of canine malignant lymphoma, where epigenetic reprogramming is being investigated as a potential treatment approach. Moreover, the study of adaptive antibiotic resistance in Escherichia coli has highlighted the role of epigenetic modifications in transient metabolic adaptations, which could inform strategies to combat antibiotic resistance in canine pathogens. By leveraging these advances and insights from other species, researchers can continue to uncover and utilize epigenetic markers to improve disease resistance and treatment outcomes in dogs (D'Aquila et al., 2023).
4 Case Study
4.1 Epigenetic profiling of disease-resistant dog breeds
Epigenetic profiling in dogs has become a crucial tool for understanding the genetic and environmental factors that contribute to disease resistance. Recent advancements in next-generation sequencing (NGS) and integrative mapping techniques have enabled researchers to generate comprehensive reference epigenomes for domesticated dogs. For instance, a study utilized transcriptome sequencing paired with histone mark and DNA methylome profiling across 11 adult tissue types to create a reference epigenome for dogs. This integrative approach allowed for the identification of distinct chromatin states and somatic super-enhancer landscapes, which are associated with various biological and disease traits. Such detailed epigenetic maps are invaluable for identifying markers linked to disease resistance in specific dog breeds (Figure 2) (Son et al., 2022).
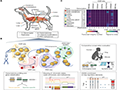 Figure 2 Overview of the integrative mapping approach to generate a dog reference epigenome (Adopted from Son et al., 2022) Image caption: (A) Diagram of 11 primary tissue types from beagle dogs sampled for the study. (B) Synopsis of next-generation sequencing (NGS) methods, data integration approaches, and analyses performed for the integrative profiling the dog epigenome. See also Methods. (C) Matrix of in-house generated NGS dataset quality from 11 primary tissue types, including information on RNA expression, defined epigenomic modifications, and DNA methylation. Normalized data integrity measures for each NGS sample profile [transcript integrity number for whole-transcriptome RNA-seq and relative strand cross-correlation coefficient (RSC) quality score for histone ChIP-seq and MBD-seq] are displayed. Two replicates per sampled tissue were profiled (Adopted from Son et al., 2022) |
4.2 Comparative analysis with disease-susceptible breeds
Comparative analysis between disease-resistant and disease-susceptible dog breeds can reveal significant differences in their epigenetic landscapes. By examining the epigenetic profiles of both groups, researchers can identify specific epigenetic markers that correlate with disease resistance or susceptibility. For example, studies in other species, such as dairy cows, have shown that single nucleotide polymorphisms (SNPs) and variations in mRNA levels of certain genes are linked to disease resistance or susceptibility. Similarly, in dogs, comparative epigenetic studies can pinpoint differences in DNA methylation patterns, histone modifications, and gene expression profiles that contribute to their ability to resist diseases.
4.3 Outcomes and implications of the case study
The outcomes of this case study have several important implications for canine health and breeding programs. Firstly, the identification of specific epigenetic markers associated with disease resistance can lead to the development of targeted therapies and preventive measures. For example, the use of histone deacetylase inhibitors and microRNAs as therapeutic agents has shown promise in treating canine lymphoma. Secondly, these findings can inform selective breeding programs aimed at enhancing disease resistance in dog populations. By incorporating epigenetic markers into breeding strategies, it is possible to produce offspring with improved resilience to diseases, thereby reducing the reliance on antibiotics and other medications (Gul et al., 2022). Lastly, the insights gained from this research can also be applied to other species, including humans, given the similarities in epigenetic mechanisms across different organisms (Essa et al., 2023).
5 Potential Applications of Epigenetic Markers
5.1 Breeding programs for enhancing disease resistance
Epigenetic markers hold significant promise for improving breeding programs aimed at enhancing disease resistance in dogs. By identifying specific DNA methylation patterns associated with disease resistance, breeders can select for these traits more effectively. This approach is akin to marker-assisted selection (MAS) used in plant breeding, where molecular markers are applied to select for disease resistance traits at an early stage. The use of epigenetic markers can similarly streamline the selection process in dogs, allowing for the early identification of individuals with desirable resistance traits. This can lead to the development of dog breeds that are more resilient to diseases, ultimately improving animal welfare and reducing the need for medical interventions (Izquierdo and Crujeiras, 2019).
5.2 Personalized veterinary medicine based on epigenetic profiles
The integration of epigenetic profiles into veterinary medicine can revolutionize the way diseases are diagnosed and treated in dogs. By analyzing the epigenetic modifications, such as DNA methylation patterns, veterinarians can gain insights into the underlying mechanisms of diseases and their progression. This personalized approach allows for the development of tailored treatment plans that consider the unique epigenetic makeup of each dog. For instance, understanding the epigenetic factors involved in insulin resistance can lead to more effective management strategies for conditions like diabetes, which are influenced by both genetic and environmental factors. Personalized veterinary medicine based on epigenetic profiles can thus enhance the precision and efficacy of treatments, improving health outcomes for dogs (Whelan et al., 2023).
5.3 Long-term health monitoring and disease prevention
Epigenetic markers offer a powerful tool for long-term health monitoring and disease prevention in dogs. By establishing baseline epigenetic profiles, it is possible to monitor changes over time and detect early signs of disease or environmental stressors. This proactive approach can facilitate early interventions, potentially preventing the onset of diseases or mitigating their impact. For example, DNA methylation clocks and environment-specific DNA methylation signatures can provide complex, context-dependent readouts about a dog's health and exposure to environmental factors. Such monitoring can be particularly valuable in managing chronic conditions and ensuring the overall well-being of dogs throughout their lives. By leveraging epigenetic markers for continuous health assessment, veterinarians can adopt a more preventive approach to animal healthcare, ultimately enhancing the quality of life for dogs (Miedaner and Korzun, 2012).
6 Challenges and Future Directions
6.1 Technical limitations in epigenetic marker discovery
The discovery of epigenetic markers for disease resistance in dogs faces several technical challenges. One significant limitation is the complexity of the canine genome and the need for high-resolution genetic mapping techniques. For instance, bulked segregant analysis (BSA) has been used successfully in plants to identify markers linked to disease resistance genes, but its application in dogs is still in its infancy. Additionally, the identification of epigenetic modifications such as DNA methylation and histone modifications requires advanced technologies like RNA sequencing (RNA-Seq) and chromatin immunoprecipitation sequencing (ChIP-Seq), which are resource-intensive and require specialized expertise. The polyploid nature of some genomes, as seen in wheat, further complicates the identification and validation of epigenetic markers, suggesting that similar complexities might be encountered in canine studies (Ramírez-González et al., 2015).
6.2 Ethical considerations in using epigenetic data for breeding
The use of epigenetic data in breeding programs raises several ethical concerns. One major issue is the potential for genetic discrimination, where dogs with certain epigenetic markers might be favored or disfavored, leading to a reduction in genetic diversity. This is particularly concerning given the role of breed-specific traits in disease prevalence and prognosis, as seen in canine malignant lymphoma. Moreover, the manipulation of epigenetic markers for breeding purposes could lead to unintended consequences, such as the exacerbation of other health issues or the loss of beneficial traits. Ethical breeding practices must therefore balance the benefits of disease resistance with the need to maintain genetic diversity and overall canine health (Bachir et al., 2022).
6.3 Future prospects for epigenetic research in canine health
Despite these challenges, the future of epigenetic research in canine health is promising. Advances in understanding the molecular mechanisms underlying diseases like insulin resistance and cancer have highlighted the potential of epigenetic markers as therapeutic targets and diagnostic tools. For example, the identification of histone deacetylases and demethylase inhibitors as potential treatments for canine lymphoma opens new avenues for targeted therapies. Additionally, the reversibility of epigenetic marks offers the possibility of developing interventions that can modify disease risk without altering the underlying genetic code4. Future research should focus on refining these techniques and exploring their applicability in canine models, ultimately aiming to improve disease resistance and overall health in dogs. By addressing these technical and ethical challenges, and leveraging the potential of epigenetic research, we can pave the way for significant advancements in canine health and disease resistance (Michelmore et al., 1991).
7 Conclusion
This study has highlighted several key findings in the identification of epigenetic markers for disease resistance in dogs. Bulked segregant analysis has been shown to be an effective method for rapidly identifying markers linked to specific genes or genomic regions, which can be applied to various species, including dogs. The role of epigenomic mechanisms, such as DNA methylation, in the onset and management of diseases like insulin resistance has been well-documented, suggesting that similar mechanisms could be at play in canine disease resistance. Furthermore, the epigenetic regulation of the ABCB1 gene in drug-sensitive and drug-resistant canine lymphoid tumor cell lines has been explored, revealing significant differences in DNA methylation and histone acetylation between these cell lines. Lastly, the review of epigenetic alterations in canine malignant lymphoma has underscored the potential of epigenetic markers as therapeutic targets and diagnostic/prognostic biomarkers.
Epigenetic markers are crucial for understanding disease resistance in dogs because they provide insights into how gene expression is regulated in response to environmental factors. These markers, such as DNA methylation and histone modifications, can influence the development and progression of diseases by altering gene expression without changing the underlying DNA sequence. Identifying these markers can lead to the development of new diagnostic tools and therapeutic strategies. For instance, the regulation of the ABCB1 gene through DNA methylation and histone acetylation has been linked to multidrug resistance in canine lymphoid tumors, highlighting the potential for epigenetic therapies to overcome drug resistance. Additionally, understanding the epigenetic mechanisms involved in diseases like insulin resistance can pave the way for preventive measures and improved management of such conditions.
Future research should focus on expanding the understanding of epigenetic mechanisms in various canine diseases and exploring their potential as therapeutic targets. Studies should aim to identify specific epigenetic markers associated with disease resistance and investigate their role in gene regulation. For example, further research into the epigenetic regulation of the ABCB1 gene could lead to the development of targeted therapies for overcoming multidrug resistance in canine cancers. Additionally, exploring the use of histone deacetylase and demethylase inhibitors as treatments for canine lymphoma could provide new avenues for therapy. The potential impact of these research directions is significant, as they could lead to more effective treatments, improved disease management, and better prognostic tools for canine diseases. By leveraging the knowledge of epigenetic markers, researchers can develop innovative strategies to enhance disease resistance and improve the overall health and well-being of dogs.
Acknowledgements
I would like to thank the anonymous reviewers for their insightful comments and suggestions that greatly improved the manuscript.
Conflict of Interest Disclosure
Author affirms that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Bachir E., Poiraud C., Claux H., Moghrabi S., Paget S., Duchêne B., Jonckheere N., Neve B., Leteurtre E., Seuningen I., and Vincent A., 2022, Abstract 2965: Epigenetic mechanisms involved in acquired resistance to combined chemotherapies in digestive cancer cells, Cancer Research, 82(12_Supplement): 2965.
https://doi.org/10.1158/1538-7445.am2022-2965
Begolli R., Sideris N., and Giakountis A., 2019, LncRNAs as chromatin regulators in cancer: from molecular function to clinical potential, Cancers, 11(10): 1524.
https://doi.org/10.3390/cancers11101524
PMID: 31658672 PMCID: PMC6826483
Ceric O., Tyson G., Goodman L., Mitchell P., Zhang Y., Prarat M., Cui J., Peak L., Scaria J., Antony L., Thomas M., Nemser S., Anderson R., Thachil A., Franklin-Guild R., Slavić D., Bommineni Y., Mohan S., Sanchez S., Wilkes R., Sahin O., Hendrix G., Lubbers B., Reed D., Jenkins T., Roy A., Paulsen D., Mani R., Olsen K., Pace L., Pulido M., Jacob M., Webb B., Dasgupta S., Patil A., Ramachandran A., Tewari D., Thirumalapura N., Kelly D., Rankin S., Lawhon S., Wu J., Burbick C., and Reimschuessel R., 2019, Enhancing the one health initiative by using whole genome sequencing to monitor antimicrobial resistance of animal pathogens: Vet-LIRN collaborative project with veterinary diagnostic laboratories in United States and Canada, BMC Veterinary Research, 15(1): 130.
https://doi.org/10.1186/s12917-019-1864-2
PMID: 31060608 PMCID: PMC6501310
D'Aquila P., Rango F., Paparazzo E., Passarino G., and Bellizzi D., 2023, Epigenetic-based regulation of transcriptome in escherichia coli adaptive antibiotic resistance, Microbiology Spectrum, 11(3): e0458322.
https://doi.org/10.1128/spectrum.04583-22
PMID: 37184386 PMCID: PMC10269836
de Vasconcelos T.C.B., Furtado M., Belo V., Morgado F., and Figueiredo F., 2017, Canine susceptibility to visceral leishmaniasis: A systematic review upon genetic aspects, considering breed factors and immunological concepts, Infection Genetics Evolution, 74: 103293.
https://doi.org/10.1016/j.meegid.2017.10.005
PMID: 28987807
Essa B., Al-Sharif M., Abdo M., Fericean L., and Ateya A., 2023, New insights on nucleotide sequence variants and mRNA levels of candidate genes assessing resistance/susceptibility to mastitis in holstein and montbéliarde dairy cows, Veterinary Sciences, 10(1): 135.
https://doi.org/10.3390/vetsci10010035
PMID: 36669036 PMCID: PMC9861242
Furtado C., Luciano M., Santos R., Furtado G., Moraes M., and Pessoa C., 2019, Epidrugs: targeting epigenetic marks in cancer treatment, Epigenetics, 14: 1164-1176.
https://doi.org/10.1080/15592294.2019.1640546
Gray J., Wani S., and Campbell M., 2022, Epigenomic alterations in cancer: mechanisms and therapeutic potential, Clinical Science, 136(7): 473-492.
https://doi.org/10.1042/CS20210449
Gul H., Habib G., Khan I., Rahman S., Khan N., Wang H., Khan N., and Liu Y., 2022, Genetic resilience in chickens against bacterial, viral and protozoal pathogens, Frontiers in Veterinary Science, 9: 1032983.
https://doi.org/10.3389/fvets.2022.1032983
PMID: 36439341 PMCID: PMC9691405
Islam M., Rony S., Rahman M., Çınar M., Villena J., Uddin M., and Kitazawa H., 2020, Improvement of disease resistance in livestock: application of immunogenomics and CRISPR/Cas9 technology, Animals, 10(12): 2236.
https://doi.org/10.3390/ani10122236
PMID: 33260762 PMCID: PMC7761152
Izquierdo A., and Crujeiras A., 2019, Role of epigenomic mechanisms in the onset and management of insulin resistance, Reviews in Endocrine and Metabolic Disorders, 20: 89-102.
https://doi.org/10.1007/s11154-019-09485-0
Ling C., and Rönn T., 2016, Epigenetic markers to further understand insulin resistance, Diabetologia, 59: 2295-2297.
https://doi.org/10.1007/s00125-016-4109-y
Mahfouz N., Ferreira I., Beisken S., Haeseler A., and Posch A., 2020, Large-scale assessment of antimicrobial resistance marker databases for genetic phenotype prediction: a systematic review, Journal of Antimicrobial Chemotherapy, 75: 3099-3108.
https://doi.org/10.1093/jac/dkaa257
Maimaiti A., Aili Y., Turhon M., Kadeer K., Aikelamu P., Wang Z., Niu W., Aisha M., Kasimu M., Wang Y., and Wang Z., 2022, Modification patterns of DNA methylation-related lncRNAs regulating genomic instability for improving the clinical outcomes and tumour microenvironment characterisation of lower-grade gliomas, Frontiers in Molecular Biosciences, 9: 844973.
https://doi.org/10.3389/fmolb.2022.844973
Michelmore R., Paran I., and Kesseli R., 1991, Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations, Proceedings of the National Academy of Sciences of the United States of America, 88: 9828-9832.
https://doi.org/10.1073/PNAS.88.21.9828
Miedaner T., and Korzun V., 2012, Marker-assisted selection for disease resistance in wheat and barley breeding, Phytopathology, 102(6): 560-566.
https://doi.org/10.1094/PHYTO-05-11-0157
Montaner-Angoiti E., Marín-García P., and Llobat L., 2023, Epigenetic alterations in canine malignant lymphoma: future and clinical outcomes, Animals, 13(3): 468.
https://doi.org/10.3390/ani13030468
PMID: 36766357 PMCID: PMC9913421
Pathania A., Prathipati P., Pandey M., Byrareddy S., Coulter D., Gupta S., and Challagundla K., 2021, The emerging role of non-coding RNAs in the epigenetic regulation of pediatric cancers, Seminars in Cancer Biology, 83: 227-241.
https://doi.org/10.1016/j.semcancer.2021.04.015
PMID: 33910063
Piletič K., and Kunej T., 2016, MicroRNA epigenetic signatures in human disease, Archives of Toxicology, 90: 2405-2419.
https://doi.org/10.1007/s00204-016-1815-7
Raei N., Safaralizadeh R., Hesseinpourfeizi M., Yazdanbod A., Pourfarzi F., and Latifi-Navid S., 2021, Crosstalk between LncRNAs and MiRNAs in gastrointestinal cancer drug resistance, Life Sciences, 284: 119933.
https://doi.org/10.1016/j.lfs.2021.119933
Ramírez-González R., Segovia V., Bird N., Fenwick P., Holdgate S., Berry S., Jack P., Cáccamo M., and Uauy C., 2015, RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat, Plant Biotechnology Journal, 13(5): 613-624.
https://doi.org/10.1111/pbi.12281
Son K., Aldonza M., Nam A., Lee K., Lee J., Shin K., Kang K., and Cho J., 2022, Integrative mapping of the dog epigenome: Reference annotation for comparative intertissue and cross-species studies, Science Advances, 9(27): eade3399.
https://doi.org/10.1101/2022.07.22.501075
Tomiyasu H., Goto-Koshino Y., Fujino Y., Ohno K., and Tsujimoto H., 2014, Epigenetic regulation of the ABCB1 gene in drug-sensitive and drug-resistant lymphoid tumour cell lines obtained from canine patients, Veterinary journal, 199(1): 103-109.
https://doi.org/10.1016/j.tvjl.2013.10.022
Tyson G., Ceric O., Guag J., Nemser S., Borenstein S., Slavić D., Lippert S., McDowell R., Krishnamurthy A., Korosec S., Friday C., Pople N., Saab M., Fairbrother J., Janelle I., McMillan D., Bommineni Y., Simon D., Mohan S., Sanchez S., Phillips A., Bartlett P., Naikare H., Watson C., Sahin O., Stinman C., Wang L., Maddox C., DeShambo V., Hendrix K., Lubelski D., Burklund A., Lubbers B., Reed D., Jenkins T., Erol E., Patel M., Locke S., Fortner J., Peak L., Balasuriya U., Mani R., Kettler N., Olsen K., Zhang S., Shen Z., Landinez M., Thornton J., Thachil A., Byrd M., Jacob M., Krogh D., Webb B., Schaan L., Patil A., Dasgupta S., Mann S., Goodman L., Franklin-Guild R., Anderson R., Mitchell P., Cronk B., Aprea M., Cui J., Jurkovic D., Prarat M., Zhang Y., Shiplett K., Campos D., Rubio J., Ramanchandran A., Talent S., Tewari D., Thirumalapura N., Kelly D., Barnhart D., Hall L., Rankin S., Dietrich J., Cole S., Scaria J., Antony L., Lawhon S., Wu J., McCoy C., Dietz K., Wolking R., Alexander T., Burbick C., and Reimschuessel R., 2021, Genomics accurately predicts antimicrobial resistance in Staphylococcus pseudintermedius collected as part of Vet-LIRN resistance monitoring, Veterinary Microbiology, 254: 109006.
https://doi.org/10.1016/j.vetmic.2021.109006
Wang X.R., Yu D.H., and Chen L., 2023, Antimicrobial resistance and mechanisms of epigenetic regulation, Frontiers in Cellular and Infection Microbiology, 13: 1199646.
https://doi.org/10.3389/fcimb.2023.1199646
PMID: 37389209 PMCID: PMC10306973
Wegener A., Broens E.M., Zomer A., Spaninks M., Wagenaar J., and Duim B., 2018, Comparative genomics of phenotypic antimicrobial resistances in methicillin-resistant Staphylococcus pseudintermedius of canine origin, Veterinary Microbiology, 225: 125-131.
https://doi.org/10.1016/j.vetmic.2018.09.013
PMID: 30322524
Whelan R., Tönges S., Böhl F., and Lyko F., 2023, Epigenetic biomarkers for animal welfare monitoring, Frontiers in Veterinary Science, 9: 1107843.
https://doi.org/10.3389/fvets.2022.1107843
PMID: 36713882 PMCID: PMC9874107
Zhou Y., Sun W., Qin Z., Guo S., Kang Y., Zeng S., and Yu L., 2020, LncRNA regulation: new frontiers in epigenetic solutions to drug chemoresistance, Biochemical Pharmacology, 189: 114228.
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Zhaolin Wang
. Xiaofang Lin
Related articles
. Canine health
. Epigenetics
. Disease resistance
. DNA methylation
. Veterinary medicine
Tools
. Post a comment


